Abundance via point-transects distance-sampling (point counts) when detection depends on covariates.
Loading required package: units
udunits database from C:/Users/trent/AppData/Local/R/win-library/4.5/units/share/udunits/udunits2.xml
data(thrasherDf)
oneHectare <- units::set_units(1, "ha")
dfuncFit <- thrasherDf |>
dfuncEstim(dist ~ bare + shrub + groupsize(groupsize)
, likelihood = "hazrate") |>
abundEstim(area = oneHectare
, ci = NULL)
summary(dfuncFit)
Call: dfuncEstim(data = thrasherDf, dist ~ bare + shrub +
groupsize(groupsize), likelihood = "hazrate")
Coefficients:
Estimate SE z p(>|z|)
(Intercept) 6.256097544 0.94375974 6.6289091 3.381767e-11
bare -0.002688512 0.01106188 -0.2430429 8.079722e-01
shrub -0.076018161 0.02869760 -2.6489382 8.074510e-03
k 4.412968498 0.43682868 10.1022865 5.396891e-24
Message: Success; Asymptotic SE's
Function: HAZRATE
Strip: 0 [m] to 265 [m]
Average effective detection radius (EDR): 121.0084 [m] (range 83.04405 [m] to 158.5773 [m])
Average probability of detection: 0.2115124 (range 0.09820311 to 0.3580883)
Scaling: g(0 [m]) = 1
Log likelihood: -994.7568
AICc: 1997.726
Surveyed Units: 120
Individuals seen: 196 in 193 groups
Average group size: 1.015544
Group size range: 1 to 2
Density in sampled area: 3.704369e-05 [1/m^2]
Abundance in 10000 [m^2] study area: 0.3704369
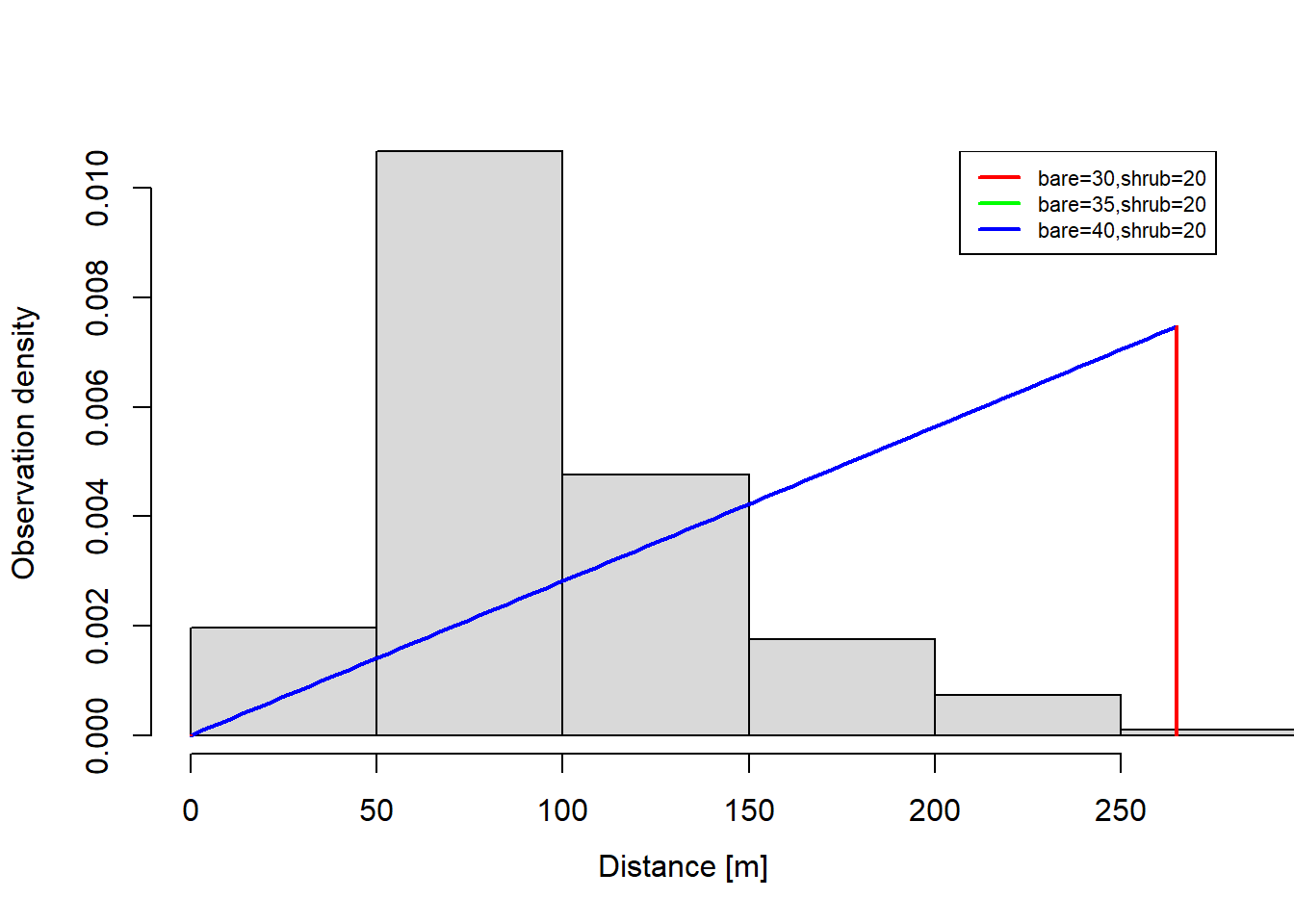
plot(dfuncFit
, newdata = data.frame(bare = c(30, 35, 40)
, shrub = 20)
, lty = 1)