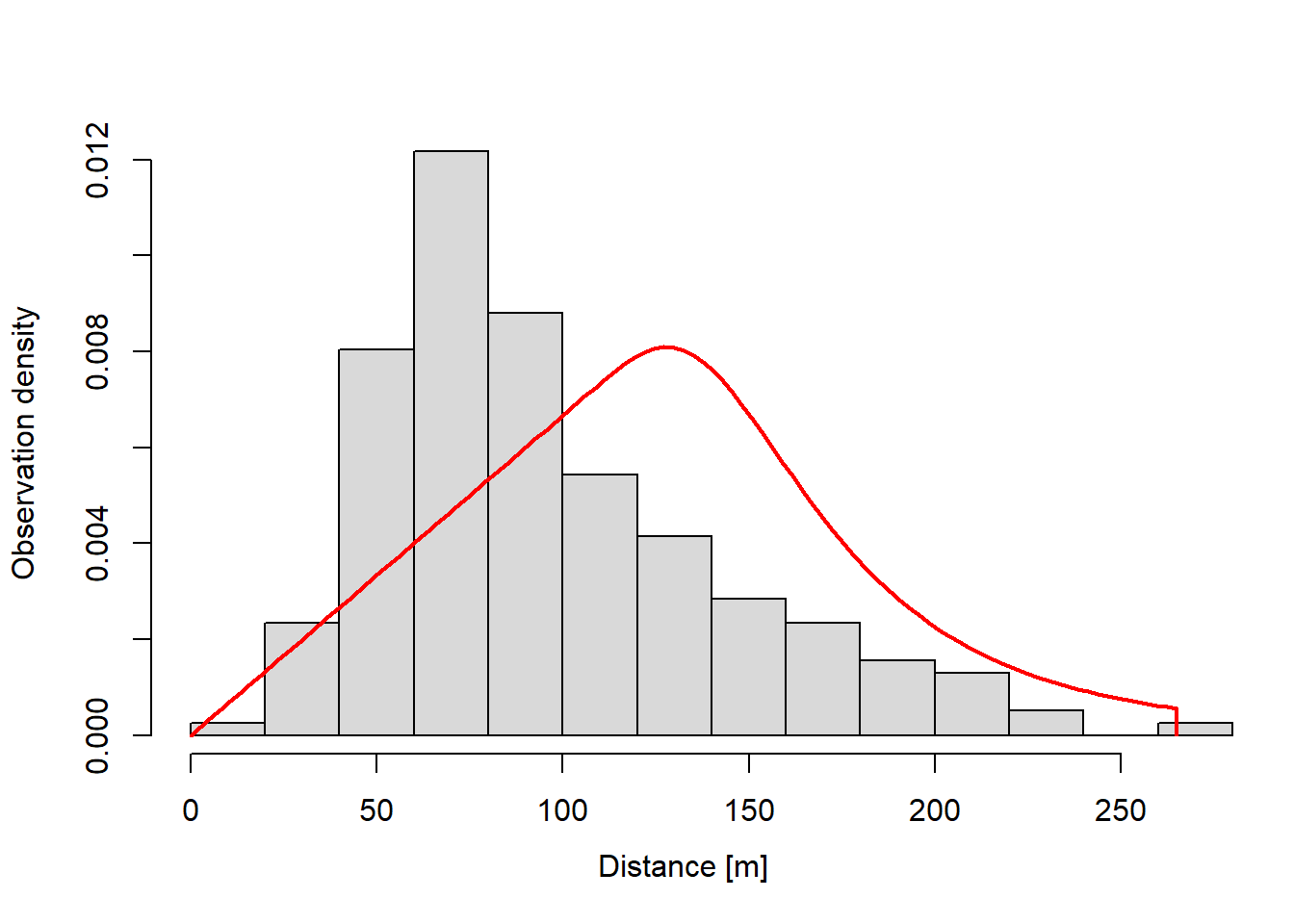
Abundance via point-transect distance-sampling (point counts) when detection does not depends on covariates.
Loading required package: units
udunits database from C:/Users/trent/AppData/Local/R/win-library/4.5/units/share/udunits/udunits2.xml
# Example data
data("thrasherDetectionData") # access example data
data("thrasherSiteData")
head(thrasherDetectionData) # inspect example data
siteID groupsize dist
1 C1X01 1 11 [m]
2 C1X01 1 183 [m]
3 C1X02 1 58 [m]
4 C1X04 1 89 [m]
5 C1X05 1 83 [m]
6 C1X06 1 95 [m]
siteID observer bare herb shrub height npoints
1 C1X01 obs5 45.8 19.5 18.7 23.7 1
2 C1X02 obs5 43.4 20.2 20.0 23.6 1
3 C1X03 obs5 44.1 18.8 19.4 23.7 1
4 C1X04 obs5 38.3 22.5 23.5 34.3 1
5 C1X05 obs5 41.5 20.5 20.6 26.8 1
6 C1X06 obs5 43.7 18.6 20.0 23.8 1
# Make nested data frame required by Rdistance
thrasherDf <- RdistDf(thrasherSiteData
, thrasherDetectionData
, by = "siteID"
, pointSurvey = TRUE
, observer = "single"
, .detectionCol = "detections"
, .effortCol = "npoints")
head(thrasherDf)
# A tibble: 6 × 8
# Rowwise: siteID
siteID detections observer bare herb shrub height npoints
<fct> <list<tibble[,2]>> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
1 C1X01 [2 × 2] obs5 45.8 19.5 18.7 23.7 1
2 C1X02 [1 × 2] obs5 43.4 20.2 20 23.6 1
3 C1X04 [1 × 2] obs5 38.3 22.5 23.5 34.3 1
4 C1X05 [1 × 2] obs5 41.5 20.5 20.6 26.8 1
5 C1X06 [1 × 2] obs5 43.7 18.6 20 23.8 1
6 C1X08 [1 × 2] obs5 41.5 20.1 20.9 29.4 1
summary(thrasherDf, formula = dist ~ groupsize(groupsize))
Transect type: point
Effort:
Transects: 120
Total length: 120 [points]
Distances:
0 [m] to 265 [m]: 193
Sightings:
Groups: 193
Individuals: 196
oneHectare <- units::set_units(1, "ha")
dfuncFit <- thrasherDf |>
dfuncEstim(dist ~ groupsize(groupsize)
, likelihood = "hazrate") |>
abundEstim(area = oneHectare
, ci = NULL)
summary(dfuncFit)
Call: dfuncEstim(data = thrasherDf, dist ~ groupsize(groupsize),
likelihood = "hazrate")
Coefficients:
Estimate SE z p(>|z|)
(Intercept) 4.540434 0.06265929 72.46226 0.000000e+00
k 4.199565 0.39721233 10.57259 3.992942e-26
Message: Success; Asymptotic SE's
Function: HAZRATE
Strip: 0 [m] to 265 [m]
Effective detection radius (EDR): 118.6254 [m]
Probability of detection: 0.2003842
Scaling: g(0 [m]) = 1
Log likelihood: -999.0241
AICc: 2002.111
Surveyed Units: 120
Individuals seen: 196 in 193 groups
Average group size: 1.015544
Group size range: 1 to 2
Density in sampled area: 3.694619e-05 [1/m^2]
Abundance in 10000 [m^2] study area: 0.3694619
plot(dfuncFit
, nbins = 15)